Original Research
2014
September
Volume : 2
Issue : 3
Analysis of HLA-A, HLA-B and HLA-DRB1 allelic frequencies in tertiary care from Telangana and Andhra Pradesh
Radhika Chowdary D, Sukrutha Gopal Reddy, Reddy VS, Pavani, Prasad Reddy k
Pdf Page Numbers :- 140-144
Radhika Chowdary D1,*, Sukrutha Gopal Reddy1, Reddy VS2, Pavani1 and Prasad Reddy K1
1Department Laboratory Medicine, Krishna Institute of Medical Sciences, Minister Road, Secunderabad - 500003, Telangana, India
2Department of Nephrology, Krishna Institute of Medical Sciences, Minister Road, Secunderabad - 500003, Telangana, India
*Corresponding author: Dr. D. Radhika chowdary, MD (Biochemistry), Consultant Biochemist & Chief of Labs, Department Laboratory Medicine, Krishna Institute of Medical Sciences, Minister Road, Secunderabad - 500003, Telangana, India. Email: rchowdary13@gmail.com
Received 17 April 2014; Revised 22 May 2014; Accepted 3 June 2014
Citation: Radhika Chowdary D, Sukrutha Gopal R, Reddy VS, Pavani, Prasad Reddy K. Analysis of HLA-A, HLA-B and HLA-DRB1 allelic frequencies in tertiary care from Telangana and Andhra Pradesh. J Med Sci Res 2014; 2(3):140-144. DOI: http://dx.doi.org/10.17727/JMSR.2014/2-025
Copyright: © 2014 Radhika Chowdary D et al. Published by KIMS Foundation and Research Center. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Introduction: The high polymorphism of the HLA system and its typification is used as valuable tool in establishing association to various illnesses, immune and genetic profiles. It also provides a guide to identify compatibility among donor and recipient of organ transplants.
Objective: To analyse HLA-A, HLA-B and HLA-DRB1 allelic frequencies among renal transplant recipients and their prospective related and unrelated donors attending Nephrology department at KIMS Hospital, Secunderabad.
Methods: 146 kidney recipients and 154 related or unrelated donors coming from different regions of Telangana and Andhra Pradesh for kidney transplantation from September 2007 to December 2013 were included in this study. All were screened for HLA-A, B and DRB, via SSP PCR based methodology and allelic frequency was established.
Result: 1738 and 17 different alleles were identified for HLA-A, B and DRB1 respectively. Alleles found more frequently are A*24, A*02, A*01, B*35, B*61, B*44 and DRB1*15, DRB1*04, DRB1*07.
Conclusion: The results obtained are useful as a reference for organ transplantations and also for allowing compatible donors within specific area for patients requiring renal transplant.
Keywords: HLA-A; HLA-B; HLA-DRB1; allelic frequencies
Full Text
Introduction
Renal transplant is the most common and standard therapy in case of renal failure. The success of transplantation largely depends on the compatibility of human leukocyte antigens (HLA) [1]. The human leukocyte antigen system (HLA) of the major histocompatibility complex (MHC) contains a numerous family of genes located on the short arm of the 6th chromosome (6p 21.1 – 6p 21.3) [2]. HLA molecule binds and presents peptide to T lymphocyte in cell mediate immune response, plays a key role in shaping the T–cell repertoire and is also associated with allograft rejection [3]. For predicting the probability of finding compatible donors in unrelated transplantation, it is valuable to have reliable estimation of HLA allele frequencies.
In this study, we carried out molecular analysis of class I HLA-A, HLA-B and class II HLA-DRB alleles by SSP-low resolution among transplant donors and recipients.
Materials and methods
This is a retrospective study conducted on renal transplant recipients and donors attending to Department of Nephrology, KIMS Hospital, Secunderabad from September 2007 to December 2013. 5-10ml of venous blood samples were collected in EDTA vials with consent from 146 renal transplant recipients and 154 donors.
HLA tying procedure
DNA from each blood sample was extracted using QIAGEN blood mini kit adhering to the protocol recommended by the manufactures. The purity and concentration of the DNA was checked by using Nanodrop 2000 spectrophotometer (Thermo scientific) and it was adjusted to >40ng/µl. HLA typing for HLA-A, B, DRB1, DRB3, DRB4 and DRB5 alleles were carried out using polymerase chain reaction-sequences specific primers (PCR-SSP) (one Lamda, Invitrogen) as per manufactures instructions on Eppendorf Gradient Thermal cycler using 96 well ABDR SSP tray. Electrophoresis was conducted on the amplification products in 2% agarose gel dye with eithidium bromide and bands were read using Syngene gel doc software. Interpretation of the result was carried out using the work sheet provided by the manufactures.
Results
17 HLA-A, 38 HLA-B and 17 HLA-DRB1 alleles were detected among recipients and donors. The most frequent alleles in the HLA –A locus were A*24 (17.71%), A*02 (15.05%), A*01 (12.95%) and A*11 (12.95) with a frequency of greater than 10%, followed by A*30, A*33 and A*28 etc. (Figures 1A and 1B).
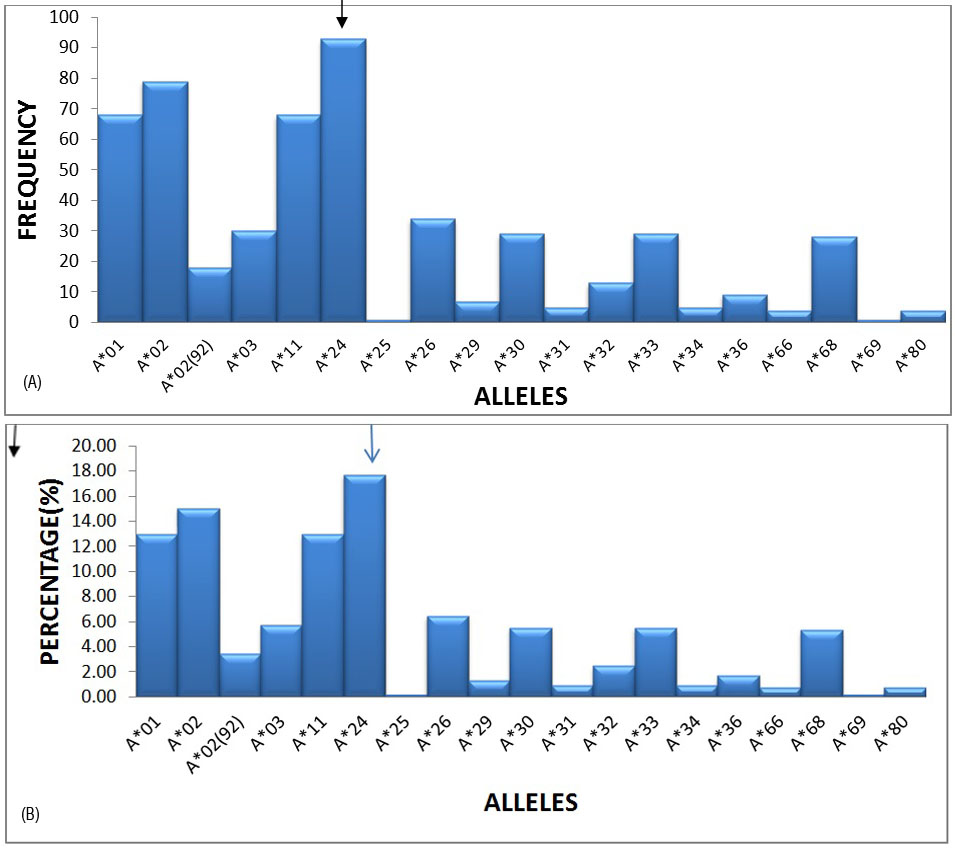
Figure 1A: Graphical representation of HLA-A locus frequencies.
Figure 1B: Graphical representation of HLA-A locus percentages.
The most frequent alleles in the HLA –B locus were B*35 (11.11%), B*51 (8.38%), B*61 (8.19%), B*44 (7.41%) and B*57 (7.41%) with a frequency of greater than 5%, followed by B*07, B*13, B*52 and B*62 etc. (Figures 2A and 2B).
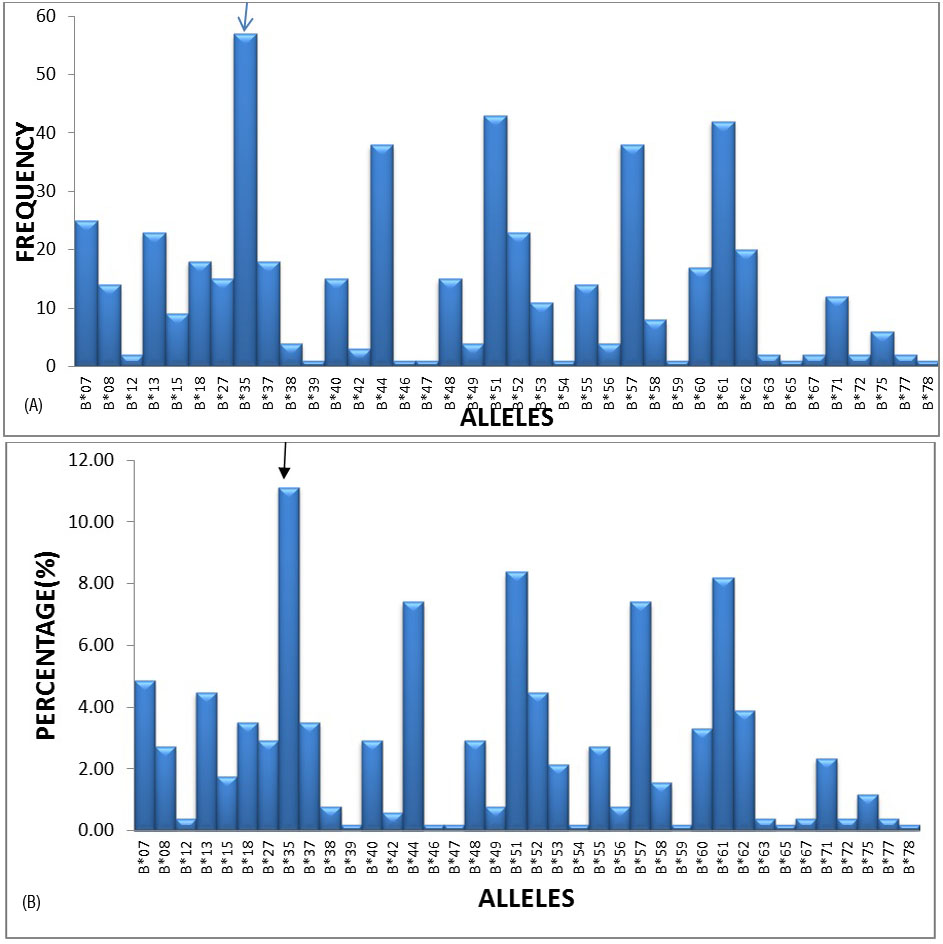
Figure 2A: Graphical representation of HLA-B locus frequencies.
Figure 2B: Graphical representation of HLA-B locus percentages.
The most frequent alleles in the HLA–DR locus were DRB1*15(DR15)(16.87%), DRB1*04(DR04)(15.86%), DRB1*07(DR07)(13.25%) and DRB1*14(DR14)(11.45%) with a frequency of greater than 10%, followed by DRB1*11(DR11), DRB1*17(DR17) and DRB1*10(DR10) etc. (Figures 3A and 3B).
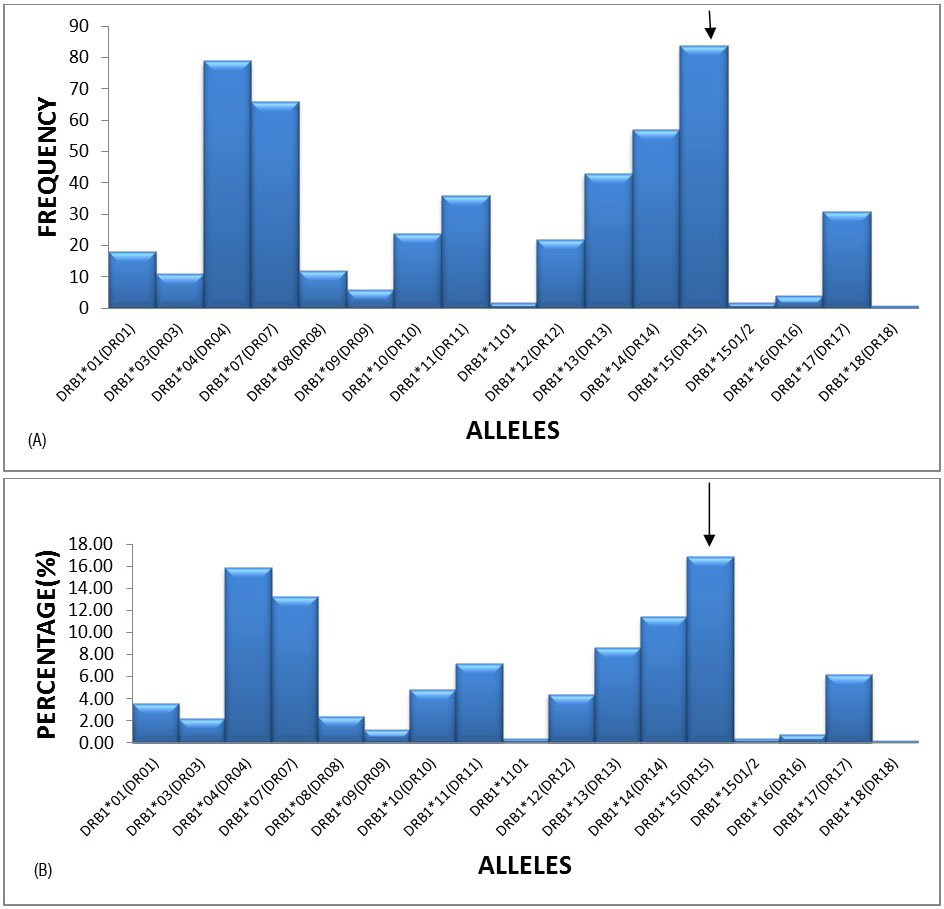
Figure 3A: Graphical representation of HLA-DRB1 locus frequencies.
Figure 3B: Graphical representation of HLA-DRB1 locus percentages.
Discussion
Analyses of HLA distribution patterns are necessary for the estimation of the likelihood of obtaining matched donors for individuals requiring transplants. Our analysis showed 17 alleles for the HLA-A, 38 for HLA-B and 17 for HLA-DRB1 whereas variable numbers of alleles were noticed in Malaysian population, 15 for HLA-A, 29 for the HLA-B and 14 for HLA-DRB1 by Dhaliwal Js et al. [4], in Colombian population 19 for HLA-A, 28 for HLA-B and 15 HLA-DRB1 by Yazmin Rocio Arias-Murillo et Al. [2] and in Nepal population 12 for HLA-A, 15 for HLA-B and 13 HLA-DRB1 by Tuladhar et al. [6].
Our analysis showed frequent HLA-A alleles as A*24 (17.71%), A*02 (15.05%) and A*01 (12.95%). Similar HLA-A allele frequency was noticed in Colombian population as A*24 (26.2%), A*02 (26%), A*01 (7.6%) by Yazmin Rocio Arias- Murillo et Al. [2], Nepal population as A*24 (17%), A*11 (34.5) by Tuladhar et Al. [6] and in Mumbai population as A*24 (15.70%), A*11 (13.17%) by S. Kankonkari et al. [7].
Similarly for HLA-B locus HLA-B*35 (11.11%) allele frequency was highest in our study population, followed by B*51 (8.38%) and B*61(8.19%), similar allele frequency were observed in Colombian population as B*35 (22.7%) and B*44(11.1%) by yazmin Rocio Arias-Murillo et al. [2], Nepal population B*33 (19%), B*35 (19%) by Tuladhar et al. [6] and in Mumbai population as B*35 (15.37%) and B*40 (12.66%) by S. Kankonkari et al. [7].
In our study regarding HLA-DRB1 alleles DRB1*15 (16.87%), DRB1* 04(15.86%) and DRB1*07(13.25%) were frequent. Similar results were observed in Colombian population DRB1*04 (24%), DRB1*15(11.3%) and DRB1*07(11.1%) by Yazmin Rocio Arias-Murillo et al. [2], Nepal population as DRB1*15(33.5%), DRB1*04(7.32%) study by Tuladhar et al. [6] and in Mumbai population as DRB1*15(19.25%), DRB1*07 (12.83%) by S. Kankonkali et al. [7].
Alleles of HLA-A and HLA-B locus, which were not detected in our analysis are A*23, A*43, A*10, A*74, B*14, B*70, B*76, B*41, B*45, B*50, B*73 and least detected alleles which were <2% on HLA-A, HLA-B and HLA –DRB1 locus are A*25, A*69, A*80, A*66, B*39, B*46, B*47, B*54, B*65, B*78, DRB1*18 and DRB1*1101. These undetected and least detected alleles were not given in other studies.
Conclusion
The present study analyzed HLA allele distribution in renal transplant recipients and donors from different regions of Telangana and Andhra Pradesh population who came for renal transplants in KIMS Hospital, Secunderabad. The results obtained are useful as a reference for organ transplantations and also for allowing compatible donors within specific area for patients requiring renal transplant.
Conflict of interest
The authors declare no conflict of interest.
References
1. Prasanavar D, Shankar Kumar. HLA: Atigen and haplotype frequencies in renal transplant recipients and donors of Maharashtra (western India). Int J Hum Gene 2004; 4:155–159.
2. Arisa-murillo YR, Bacteriol, Castro-Jiménez MA, Ríos-Espinosa MF, López-Rivera JJ et al. Analysis of HLA-A,HLA-B, HLA-DRB1 allelic genotypic and haplotypic frequencies in Colombian population. Colombia Medica 2010; 41(4):336–343.
3. Alper CA, Larsen CE, Dubey DP, Awdeh ZL, Fici DA, et al. The haplotype structure of the human major histocompatibility complex. Hum immunol 2006; 67:73–84.
4. Dhaliwal JS1, Shahnaz M, Too CL, Azrena A, Maiselamah L, et al. HLA-A, -B and -DR allele and haplotype frequencies in Malays. Asian Pac J Allergy Immunol. 2007; 25(1):47–51.
5. Burt C, Cryer C, Fuggle S, Little AM, Dyer P. HLA-A, -B, -DR allele group frequencies in 7007 kidney transplant list patients in 27 UK centres. Int J Immunogenet. 2013; 40:209–215, do; 10:1111/iJi:12000.
6. Tuladhar A, Shrestha S, Raut PP, Bhandari P, Shrestha P. HLA antigen distribution in renal transplant recipients and donors. J Nepal Health Res Counc. 2013; 11(25):289–92.
7. Kankonkari S, Shankar Kumar U. Molecular Diversity of HLA-A, HLA-B, HLA-DRB1 and HLA-DQB1 Alleles from Mumbai India: Int J Hum Genet. 2012; 12(1):57–62.
8. Bardi MS1, Jarduli LR, Jorge AJ, Camargo RB, Carneiro FP, et al. HLA-A, B and DRB1 allele and haplotype frequencies in volunteer bone marrow donors from the north of Parana State. Rev Bras Hematol Hemoter. 2012; 34(1):25–30. doi: 10.5581/1516-8484.20120010.